Gene set – disease relationship analysis in GeneAnalytics leverages the information available in MalaCards, the human disease database. MalaCards unifies and integrates data from more than 60 sources, providing a comprehensive coverage of known diseases, including disease categorizations and disease-associated gene aggregation, along with gene-disease association rankings and annotations leveraging GeneCards.
Expression & function based, disease association analysis by GeneAnalytics enables you to:
Gene- disease associations in MalaCards are divided into the following categories:
Learn more about MalaCards and its data sources.
The disease matching score is determined by the following parameters:
As described above, there are three types of relation:
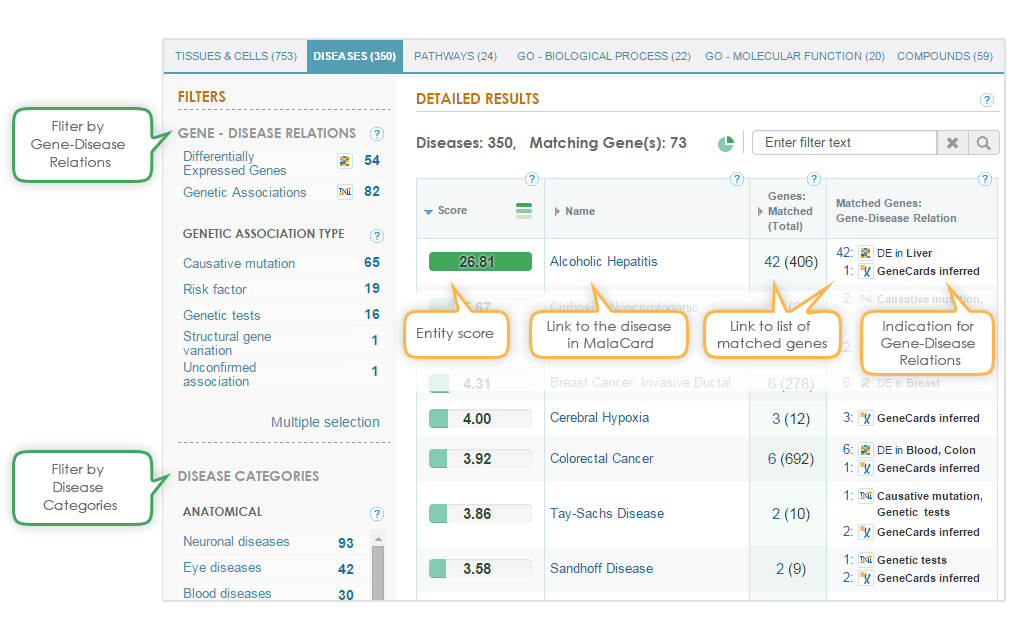
The matched diseases can be filtered either by the gene-disease relation or by MalaCards categories (see below). The gene-disease relation filter filters results to present only diseases that have either known differentially expressed genes or genes with genetic association to the disease.
Importantly, there are different types of genetic associations, which can be used to further filter the results, using the “genetic association type” filter.
Read more about Genetic association types.
MalaCards categorizes diseases into, global (fetal, genetic, cancer and infectious) and anatomical (e.g., eye, ear, liver, blood) diseases. A disease can be associated with several anatomical and global categories and is categorized as follows: