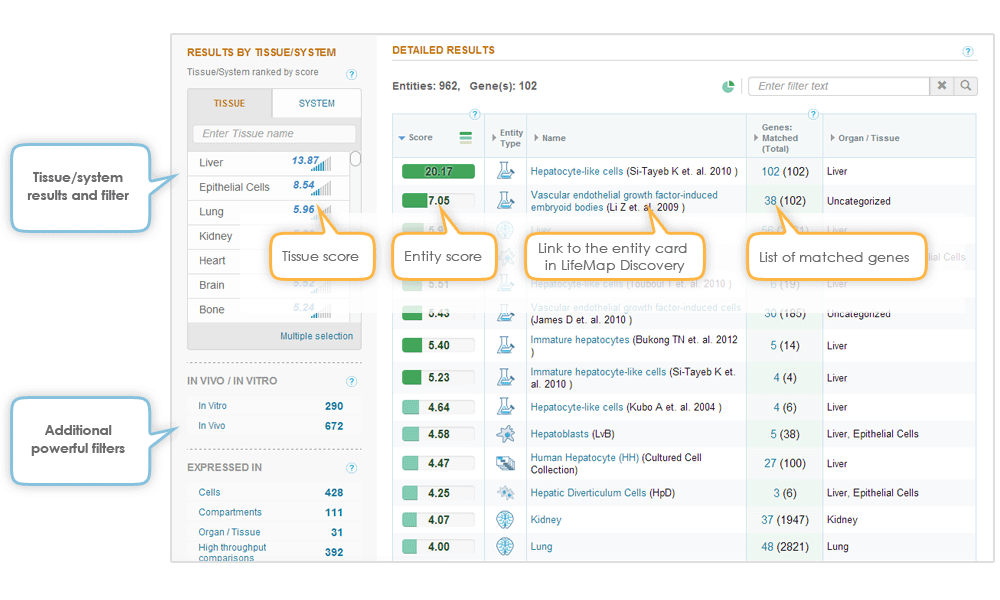
The Tissues & Cells section in GeneAnalytics leverages the extensive expression data available in LifeMap Discovery® and provides detailed information about normal cells, anatomical compartments, organs, tissues and high throughput experiments whose reported gene expression profiles match your gene set. The results are also classified into tissues and systems.
Expression-based, Tissues & Cells analysis by GeneAnalytics can help you:
Gene expression data of disease-related tissues and cells can be viewed in the disease section
Expression-based, Tissues & Cells analysis in GeneAnalytics relies on data that were manually collected from the scientific literature as well as data derived from high throughput experiments and further modeled, annotated and integrated into LifeMap Discovery.
The gene expression data is filtered to remove data lacking sufficient supporting evidence. In addition, data from mutant animals or from patients are not included in this section results, in efforts to eliminate incorrect association of aberrant gene expression profiles with normal tissues and cells.
The data available in LifeMap Discovery are obtained from the following sources (for a detailed explanation and list of sources please click here):
The following entities in LifeMap Discovery include gene expression information and are analyzed by GeneAnalytics for genes matching your gene set:
| Entity type | Available data | Notes | Example |
|---|---|---|---|
|
These entities contain list of genes that have been found to be expressed in samples taken from the whole tissue. | Heart | |
|
These entities describe specific temporospatial regions within an organ/tissue. | Renal Collecting Duct System | |
|
Inner Cell Mass Cells (ICM) Trabecular meshwork-derived mesenchymal stem cells | ||
|
These entities contain the gene list for each sample of an LSDS (see above for explanation about LSDSs) These entities serve as a match for GeneAnalytics only if their gene list is not contained within any of the above card types. |
GUDMAP: Ovary |
The matching analysis in GeneAnalytics is based on gene annotations available in LifeMap Discovery for each gene in each entity. These annotations are based on information reported in the scientific literature and/or on bioinformatics calculations executed on expression data in LifeMap Discovery.
Each gene can have one or more of the following annotations:
To receive a list of all genes expressed in a specific tissue, organ or developmental path, including annotations for selective markers, specific genes and tissue-enriched genes, please contact us.
![]()