Powered by LifeMap's GeneCards Suite integrated knowledgebase, which utilizes data from >120 select sources:
Expression-based analysis is based on data which were manually collected, filtered, modeled, annotated and integrated in our knowledgebase.
Gene expression data for normal and diseased tissues and cells are separated and displayed in different sections.
Provides the most valuable results without the need for complex bioinformatics expertise or tools. Developed by biologists, for biologists!
Results are directly linked to detailed cards in the LifeMap integrated biomedical knowledgebase and to relevant external data sources.
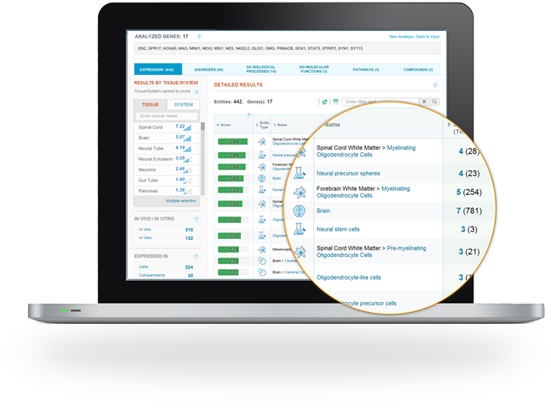
Provides categorized results lists of matched tissues, cells, diseases, pathways, compounds and gene ontology (GO) terms to enhance gene set interpretation.
The expression-based matching algorithm considers gene annotations including gene -disease association and gene specificity, enrichment or abundance in each specific tissue or cell.
GeneAnalytics enables researchers to identify tissues and cell types related to their gene sets, to characterize tissue samples and cultured cells and assess their purity and explore their selective markers.
The key strength of GeneAnalytics stems from the extensive manually curated gene expression data available in LifeMap Discovery.
Learn more >
GeneAnalytics enables researchers to identify diseases related to their gene sets, and to discover disease mechanisms and specific disease markers.
GeneAnalytics disease-related outputs rely on the information available in MalaCards, the most comprehensive human disease database.
Learn more >
GeneAnalytics enables researchers to identify pathways related to their gene sets, to explore pathway mechanisms and to define gene roles within a pathway.
GeneAnalytics pathway-related outputs rely on the information available in PathCards, the only resource to integrate data from multiple pathway resources into super-pathways.
Learn more >
GeneAnalytics enables researchers to identify Gene Ontology (GO) terms related to their gene sets, providing information about the molecular functions and biological roles of the genes of interest.
GeneAnalytics exploits the information available in the Gene Ontology (GO) project, and integrated in GeneCards – the human gene database.
Learn more >
GeneAnalytics enables researchers to identify compounds related to their gene sets, and further link to biochemical and pharmacological information about drugs, small molecules and metabolites, their mechanisms of action and their targets.
GeneAnalytics takes advantage of multiple sources of information related to more than 83,000 compounds, including those found in GeneCards, the human gene database.
Learn more >
"...I like GeneAnalytics for its convenience and accessibility of further information about specific terms, backed up by the excellent knowledgebase of GeneCards. The interface definitely helps to understand the biology faster and deeper".
“I find GeneAnalytics a powerful tool that is extremely helpful in analyzing next generation sequencing data as it allows the fast and efficient analysis of gene sets by integrating data from a number of important databases ...”
“GeneAnalytics provides complete information about genes of interest as well as precise analysis of how these genes are connected to different tissues, diseases, function and drugs. The interface is very user friendly...”
Ben-Ari Fuchs S, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, Bogoch Y, Plaschkes I, Shitrit A, Rappaport N, Kohn A, Edgar R, Shenhav L, Safran M, Lancet D, Guan-Golan Y, Warshawsky D, and Strichman R. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data, OMICS(2016) [PDF]